Biological Chemistry
Structural Biology and Biophysics by NMR
Professor Gottfried Otting
 The group develops novel tools for biomolecular applications
of NMR spectroscopy. Emphasis is placed on extending the range of
protein targets that can be investigated by NMR in pharmaceutical drug
development. Thus, methods are developed for rapid identification and
characterization of ligand binding sites, including protein-protein
and protein-DNA interactions. In addition, NMR is used to determine
the three-dimensional structures of proteins and protein domains. This
research is supported by an 800 MHz NMR spectrometer to be installed
at the beginning of 2004.
The group develops novel tools for biomolecular applications
of NMR spectroscopy. Emphasis is placed on extending the range of
protein targets that can be investigated by NMR in pharmaceutical drug
development. Thus, methods are developed for rapid identification and
characterization of ligand binding sites, including protein-protein
and protein-DNA interactions. In addition, NMR is used to determine
the three-dimensional structures of proteins and protein domains. This
research is supported by an 800 MHz NMR spectrometer to be installed
at the beginning of 2004.
We recently discovered that binding of a paramagnetic lanthanide
ion at a specific protein site provides a novel route to assign the
NMR signals of the protein to its specific amino acids with
unprecedented ease and speed.
To follow up on this discovery, we are currently working on widely
applicable methods to attach lanthanide ions to proteins that
don’t have a natural ion binding site. One such strategy
involves the synthesis of chemical compounds which on one side
specifically attach to cysteines in proteins and on the other side
carry a paramagnetic lanthanide ion. The work includes the production
of proteins containing single cysteines at specific sites and the
application of high-yield in vitro protein expression
techniques which are developed in collaboration with Dr N.E. Dixon to
allow inexpensive residue-selective 15N-labelling of
proteins.
Labelling proteins with lanthanide tags opens up a wide range of
applications which where hitherto difficult or impossible to address
by NMR or other methods. For example, they will provide a tool for 3D
structure determination of small regions in large proteins,
i.e. to “zoom” in on a region of a protein and
study its structure without having to analyze the rest of the
protein. It has long been known that lanthanides provide structural
information to NMR spectroscopists. The lanthanide tagging approach
promises to broaden these applications considerably. For example, it
will provide information about the orientation of small chemicals
(drug candidates) as they bind to protein targets. Finally, lanthanide
labelling will allow the characterization of large amplitude motions
of proteins with unprecedented accuracy.
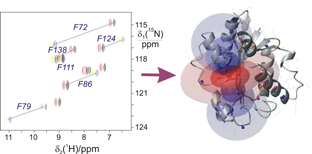 From NMR spectra to resonance assignment, structure
determination and interaction studies of proteins. A wealth of
structural information is gained by tagging with a
paramagnetic ion. Isosurfaces of the anisotropic magnetic
susceptibility (blue and red) are superimposed on the
structure of the N-terminal domain of the proofreading
exonuclease epsilon.
From NMR spectra to resonance assignment, structure
determination and interaction studies of proteins. A wealth of
structural information is gained by tagging with a
paramagnetic ion. Isosurfaces of the anisotropic magnetic
susceptibility (blue and red) are superimposed on the
structure of the N-terminal domain of the proofreading
exonuclease epsilon.
Highlights of the year were the development of an algorithm to
determine sequence-specific resonance assignments of selectively
stable-isotope labelled and lanthanide-tagged proteins by comparison
with data predicted from the 3D structure of the protein, a study of
the residence times of water molecules on protein surfaces which
reconciles NMR results with the results from molecular dynamics
calculations, and the completion of a 3D structure determination for
CLP.
Professor Gottfried Otting continues to supervise his former
laboratory at the Karolinska Institute in Stockholm. Continuing major
collaborations are with Dr Nicholas Dixon and Dr Max Keniry
(in-house), Dr Thomas Huber (Queensland University) Dr Edvards
Liepinsh (Karolinska Institute), Dr Anatoly Sharipo (Latvian
University), Dr Laszlo Patthy (Hungarian Academy of Sciences) and an
EU network on cross-correlation effects in NMR led by Professor
Geoffrey Bodenhausen (Paris).
New Algorithm for Assignment of NMR Spectra
The assignment of NMR resonances to specific protons of a protein
is a time-consuming task which can be very much shortened by the use
of a novel strategy, if the three-dimensional structure of the protein
is known and a lanthanide ion can be bound to the protein at a
specific site. The strategy has been verified for a 30 kDa
15N-labelled complex between the E. coli proteins
epsilon and theta.
(with N. Dixon, M. Keniry, A. Park, and T. Huber [U. Queensland],
G. Pintacuda [Karolinska Institute, Stockholm])
In vitro Expression of Residue-Selectively Isotope Labelled
Samples
The cell-free expression system available in Dr Nicholas
Dixon’s laboratory was used to express samples of selectively
15N labelled human cyclophilin. The yields were
sufficiently high that NMR spectra (15N-HSQC spectra) could
be recorded straight from the reaction medium without any protein
purification or concentration step. The spectra were analysed for
metabolic side reactions of the labelled amino acids that might be
catalyzed by enzymes present in the reaction medium. The data provide
a catalogue of spurious signals which can be encountered in NMR
spectra of in vitro synthesized and unpurified protein
samples.
(with N.E. Dixon, K. Ozawa)
Homonuclear CSA/DD Cross-Correlated Relaxation in COSY
The cross-correlated relaxation between the chemical shift
anisotropy (CSA) of amide protons and the dipolar field from the
α protons in the same amino acid was investigated. Experimental
results disagree with predictions from DFT simulations in
vacuo, indicating that solvation significantly affects the amide
proton CSA tensor.
(with P. Wu)
Protein Solvation by NMR and MRD
The residence time of hydration water molecules on the surface of
proteins and peptides was investigated by a high-resolution NMR
spectroscopy and magnetic resonance dispersion (MRD). A new relaxation
model assuming different diffusion coefficients of hydration and bulk
water provides a consistent theory which explains the data obtained
with both techniques. The result shows that solvent-exposed hydration
water molecules have residence times in the picosecond time range even
at temperatures near the freezing point of water.
(with B. Halle, K. Modig [Lund U., Sweden], E. Liepinsh
[Karolinska Institute, Stockholm])
Protein Structure Determinations
The 3D structure of human CLP was completed. The protein binds to
5-lipoxygenase which is an important drug target for the
suppression of inflammation.
(with E. Liepinsh, O. Rådmark [Karolinska Institute, Stockholm])
[
Otting Group |
RSC Annual Report Index ]
Last revised 18 April 2004 -
Please direct all enquiries to:
Research School of Chemistry
Authorised by the Dean, RSC as relevant officer.
©
2004 The Australian National University
CRICOS Provider Number 00120C
 From NMR spectra to resonance assignment, structure
determination and interaction studies of proteins. A wealth of
structural information is gained by tagging with a
paramagnetic ion. Isosurfaces of the anisotropic magnetic
susceptibility (blue and red) are superimposed on the
structure of the N-terminal domain of the proofreading
exonuclease epsilon.
From NMR spectra to resonance assignment, structure
determination and interaction studies of proteins. A wealth of
structural information is gained by tagging with a
paramagnetic ion. Isosurfaces of the anisotropic magnetic
susceptibility (blue and red) are superimposed on the
structure of the N-terminal domain of the proofreading
exonuclease epsilon.
 The group develops novel tools for biomolecular applications
of NMR spectroscopy. Emphasis is placed on extending the range of
protein targets that can be investigated by NMR in pharmaceutical drug
development. Thus, methods are developed for rapid identification and
characterization of ligand binding sites, including protein-protein
and protein-DNA interactions. In addition, NMR is used to determine
the three-dimensional structures of proteins and protein domains. This
research is supported by an 800 MHz NMR spectrometer to be installed
at the beginning of 2004.
The group develops novel tools for biomolecular applications
of NMR spectroscopy. Emphasis is placed on extending the range of
protein targets that can be investigated by NMR in pharmaceutical drug
development. Thus, methods are developed for rapid identification and
characterization of ligand binding sites, including protein-protein
and protein-DNA interactions. In addition, NMR is used to determine
the three-dimensional structures of proteins and protein domains. This
research is supported by an 800 MHz NMR spectrometer to be installed
at the beginning of 2004.