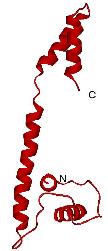
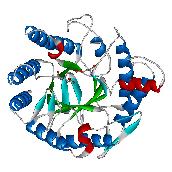
Glutathione S-transferases from Mosquitos
In collaboration with workers at Mahidol University in Thailand, we
are investigating
the structure and function of glutathione S-transferases (GST) from
the mosquito Anopheles dirus species B, an important malaria
vector in South-East Asia. These enzymes are important, because they
can break down pesticides used to control mosquitos. We have so far
determined the structure of two isozymes from an unusual gene that
gives variants through alternate splicing. In the long term, we aim
to understand how the enzymes bind and detoxify pesticides and how
this might be ameliorated.
|

A glutathione S-transferase from Anopheles dirus
|
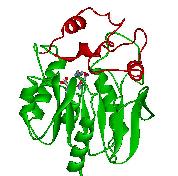 Haloalkane dehalogenase LinB
Haloalkane dehalogenase LinB
 Structural
biology lies at the nexus between chemistry, biology, and medicine.
The three dimensional structures of biological molecules such as
proteins and DNA yield a great deal of information about how they
work, and can give rise to new hypotheses about its function that can
be further probed through mutagenesis, kinetic and other biochemical
analyses. Understanding the structure and function of proteins is of
primary importance to medicine, biochemistry, and molecular genetics,
since proteins drive and regulate these processes. Protein
crystallography is our method of choice for structure determination
efforts. We gather additional information about proteins from
computational approaches such as molecular dynamics. We are
investigating the structure and function of several proteins:
Structural
biology lies at the nexus between chemistry, biology, and medicine.
The three dimensional structures of biological molecules such as
proteins and DNA yield a great deal of information about how they
work, and can give rise to new hypotheses about its function that can
be further probed through mutagenesis, kinetic and other biochemical
analyses. Understanding the structure and function of proteins is of
primary importance to medicine, biochemistry, and molecular genetics,
since proteins drive and regulate these processes. Protein
crystallography is our method of choice for structure determination
efforts. We gather additional information about proteins from
computational approaches such as molecular dynamics. We are
investigating the structure and function of several proteins: