Biological Chemistry

Spermine, an aliphatic polycationic molecule found in all cells, has an essential role in cell growth and differentiation. At present, there is no thorough understanding of how polyamines exert their physiological effects. Spermine is known to interact both with DNA and with proteins, yet the details of these interactions and the molecular basis of the biological function of spermine are poorly understood. There is evidence in the literature that spermine interacts with different forms of DNA in distinct and divergent modes. We have confirmed this and have characterised the complexes of spermine with duplex B-DNA and G-DNA using a specifically 13C-labelled spermine and advanced NMR techniques to take advantage of the specific isotope label on spermine. 13C T1 and T2 relaxation times and homonuclear and heteronuclear NOEs are used to characterise the dynamics of spermine in the presence of different forms of DNA. Spermine is bound more tightly to folded forms of G-DNA than B-DNA or linear G-DNA. Work is in progress to determine detailed dynamical models of the interaction of spermine with DNA to expand this work to the effect of spermine on protein-DNA complexes. (with J. Coughlan).
Calothrixin A and B are novel pentacyclic metabolites from cyanobacteria that exert growth-inhibitory effects at nanomolar concentrations against rapidly proliferating cell cultures. The binding properties of the calothrixins and their synthetic analogues with various structural forms of DNA are under investigation by NMR, circular dichroism and fluorescence. (with E.A. Owen, R. Rickards, and C. Chai, M. Waring [Chemistry, ANU], G.D. Smith [BAMBI, ANU])
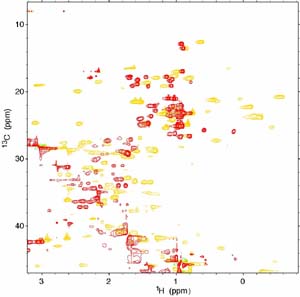
The catalytic core of Escherichia coli DNA polymerase III contains three tightly associated subunits (α, ε and θ). The refinement of the three-dimensional structure of the θ subunit was completed by the NMR group. The θ subunit has three α-helices in the N-terminal two thirds of the protein that fold to form a triangular shape. As part of a program aimed at understanding the molecular mechanism of the core, we have set out to investigate the association of the θ and ε subunits. The structure of the θ subunit bound to ε has been refined using an innovative technique that combines NOE restraints with distance and orientation restraints calculated from a paramagnetic centre located in the active site of ε. The basic structure of θ has not changed but two of the helices that were poorly defined in the uncomplexed θ subunit are properly formed in the complex. We have recently mapped the binding surface of ε on θ using advanced NMR techniques. Not surprisingly, the surface corresponds to a hydrophobic patch on θ formed on one of the previously ill-formed α-helices. Work is now in progress to align θ with ε using the same paramagnetic restraints that were used to refine the structure of θ. (with E.M. Bulloch, N.E. Dixon, S. Hamdan, T.K. Ronson, G. Otting, G. Pintacuda, and S.E. Brown [Entomology, CSIRO])

13C-HSQC NMR spectrum of the θε
complex showing unshifted peaks (red) and paramagnetic
shifted peaks
(gold) due to a lanthanide ion located at the ε
active site.
ESX is a protein that belongs to the Ets family of transcription factors. Ets proteins exhibit diverse roles in development, cell differentiation and tissue-specific gene expression and are implicated in cancers such as acute myeloid leukemia and Ewings sarcoma. The ESX transcription factor may have a role in the activation of the HER2/neu oncogene, which is overexpressed in over 40% of breast tumours. We are interested in determining the structure of ESX using X-ray crystallography and NMR. To this end we have overexpressed the C-terminal end of ESX containing the two DNA-binding domains. Attempts will be made to crystallize this fragment. A 15N-labelled form of the C-terminal fragment of ESX is also available and NMR studies will commence shortly. The long-term goal is to determine the complete structure of ESX and studies to optimize expression and folding of the N-terminal fragment and the complete protein are envisaged. This project is supported in part by a Yamagiwa–Yoshida travel grant from the International Union against Cancer. (with C.C. Benz, G. Scott [Buck Institute for Age Research, USA])