Biological Chemistry
Protein Synthesis and Evolution
Dr Nicholas Dixon
 Some proteins are enzymes that promote chemical reactions;
others provide molecular switches that control metabolic and
developmental processes through precise interactions with other
proteins, nucleic acids and other ligands. In two distinct research
programs, we are exploring the chemistry that governs the specificity
and strength of interactions of proteins with other proteins, and
ligands like substrates, inhibitors and nucleic acids.
Some proteins are enzymes that promote chemical reactions;
others provide molecular switches that control metabolic and
developmental processes through precise interactions with other
proteins, nucleic acids and other ligands. In two distinct research
programs, we are exploring the chemistry that governs the specificity
and strength of interactions of proteins with other proteins, and
ligands like substrates, inhibitors and nucleic acids.
The first program concerns the thirty or so different proteins that
collaborate to replicate the DNA of the bacterial chromosome prior to
cell division. DNA replication commends itself as a model system to
study general aspects of protein–protein and
protein–nucleic acid interactions because the proteins act
together in a giant nucleoprotein assembly called the replisome, to
make perfect copies of the chromosome. We use molecular genetics to
engineer rich sources of the proteins and to produce mutant
derivatives and segments of them, and conventional enzymology, DNA
synthesis assays and protein chemistry to study protein
function. Protein X-ray crystallography, ESR and high-field NMR
spectroscopy, mass spectrometry, electron microscopy and computational
methods are used with collaborating laboratories to further understand
the structure of the individual proteins, and to relate their
structures to how they work and interact with each other and DNA. This
year, we have focussed our efforts on interactions between the
replicative helicase DnaB and the DnaG primase, and on the ten
different subunits of DNA polymerase III (Pol III) holoenzyme, the
enzyme that actually synthesises new DNA chains during chromosomal DNA
replication.
 Our other research program has complementary objectives. A suite of
new techniques in protein chemistry is being developed, including
methods for in vitro evolution of new protein functions, in
vitro synthesis of proteins on a preparative scale, library
methods for precise location of boundaries between distinct folded
domains in larger proteins, and stabilisation of small protein domains
by end-to-end cyclisation of their polypeptide chains. Used together,
these techniques are helping to overcome some of the major bottlenecks
in rapid determination of protein structures and functions, thereby
increasing the efficiency of worldwide efforts in structural and
functional genomics. They are also being used to study fundamental
aspects of the relationship between the structure, folding, stability
and functions of proteins.
Our other research program has complementary objectives. A suite of
new techniques in protein chemistry is being developed, including
methods for in vitro evolution of new protein functions, in
vitro synthesis of proteins on a preparative scale, library
methods for precise location of boundaries between distinct folded
domains in larger proteins, and stabilisation of small protein domains
by end-to-end cyclisation of their polypeptide chains. Used together,
these techniques are helping to overcome some of the major bottlenecks
in rapid determination of protein structures and functions, thereby
increasing the efficiency of worldwide efforts in structural and
functional genomics. They are also being used to study fundamental
aspects of the relationship between the structure, folding, stability
and functions of proteins.
DNA Polymerase III Holoenzyme
The
replisome is made up of several molecular machines that interact
physically with each other. One is Pol III, the replicative DNA
polymerase, which consists of the catalytic core (α,
ε and θ
subunits), the sliding clamp (Β2),
and the six subunits of the clamp loader (δ,
δ', γ,
τ, &chi
and &upsilon). This year, methods have
been developed for purification of large quantities of the α
(polymerase) subunit and the four largest subunits of the clamp
loader, and complexes of them have been isolated for structural and
functional studies. In collaboration with a group in CSIRO, these
proteins have been used to further understand the way that the α
and δ subunits compete for their
interaction site on the β2
sliding clamp. In 2002, we reported the high-resolution crystal
structure of the ε (proofreading
exonuclease) subunit, in a complex with a nucleotide inhibitor and
two Mn(II) ions. This year, structures of complexes with other
nucleotides and metal ions have been refined, and a method has been
devised for making crystals of the apoenzyme. This sets the stage for
further studies of the mechanism of action of this important
binuclear metallohydrolase. Intein-mediated protein cyclisation has
been used to produce circularised forms of ε
that are more stable than the native form, and their structures have
also been determined. The structure of the θ
subunit in its complex with ε has
been determined by NMR spectroscopy, and a new method using
lanthanide derivatives of ε was
used to produce a model for the structure of the ε-θ
complex.
(with P.D. Carr, S. Jergic, M.A. Keniry, P.E. Lilley, D.L. Ollis,
G. Otting, K. Ozawa, A.Y. Park, P. Prosselkov and C.M. Elvin,
G. Wijffels [CSIRO, Brisbane], B. Hankamer [U. Qld], G. Pintacuda
[Karolinska Institutet, Stockholm])
The DnaB Helicase
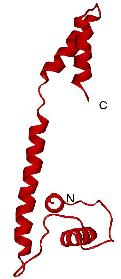 The DnaB interaction domain of DnaG primase.
The DnaB interaction domain of DnaG primase.
|
The molecular motor that drives the replisome and separates the two
strands of DNA at the apex of the replication fork is the ring-shaped
hexameric DnaB helicase. This year, we have concentrated on examining
the defects in function of a series of designed mutants of DnaB, and
on its interaction with the replicative priming enzyme, DnaG primase.
We have shown that the small C-terminal domain of primase
(DnaG-C) contains all of the determinants for interaction with DnaB,
and in collaboration with a group at the University of Western
Australia have solved the crystal structure of DnaG-C. The protein is
unusual in that it is comprised of two small helical domains linked by
a long naked helix. NMR studies were used to confirm that the
structure in solution is the same as in the crystal.
(with B. Bancia, P.E. Lilley, K.V. Loscha, G. Otting,
P.M. Schaeffer, N.K. Williams and J.M. Carazo, Y. Robledo [Centro
Nacional de Biotecnología, Madrid], J.M. Guss [U. Sydney],
E. Liepinsh [Karolinska Institutet, Stockholm], A.J. Oakley,
M.C.J. Wilce [U. Western Australia])
|
New Protein Technologies
Substantial
progress has been made in development of new methods for directed
molecular evolution of proteins with new binding specificities, for
intein-mediated end-to-end cyclisation of protein domains and
peptides, for preparative in vitro protein synthesis, for
site-specific incorporation of unnatural amino acids and lanthanide
chelates into proteins, and for the use of library methods for
protein domain identification.
(with M. Headlam, P.E. Lilley, M. Mulcair, G. Otting, K. Ozawa, P.
Prosselkov, P.M. Schaeffer, N.K. Williams and R. Dean [U. Canberra],
M. Ehrenberg [U. Uppsala, Sweden], J.L. Beck, M.M. Sheil [U.
Wollongong], J.M. Matthews [U. Sydney])
[
Dixon Group |
RSC Annual Report Index ]
Last revised 17 April 2004 -
Please direct all enquiries to:
Research School of Chemistry
Authorised by the Dean, RSC as relevant officer.
©
2004 The Australian National University
CRICOS Provider Number 00120C
 Our other research program has complementary objectives. A suite of
new techniques in protein chemistry is being developed, including
methods for in vitro evolution of new protein functions, in
vitro synthesis of proteins on a preparative scale, library
methods for precise location of boundaries between distinct folded
domains in larger proteins, and stabilisation of small protein domains
by end-to-end cyclisation of their polypeptide chains. Used together,
these techniques are helping to overcome some of the major bottlenecks
in rapid determination of protein structures and functions, thereby
increasing the efficiency of worldwide efforts in structural and
functional genomics. They are also being used to study fundamental
aspects of the relationship between the structure, folding, stability
and functions of proteins.
Our other research program has complementary objectives. A suite of
new techniques in protein chemistry is being developed, including
methods for in vitro evolution of new protein functions, in
vitro synthesis of proteins on a preparative scale, library
methods for precise location of boundaries between distinct folded
domains in larger proteins, and stabilisation of small protein domains
by end-to-end cyclisation of their polypeptide chains. Used together,
these techniques are helping to overcome some of the major bottlenecks
in rapid determination of protein structures and functions, thereby
increasing the efficiency of worldwide efforts in structural and
functional genomics. They are also being used to study fundamental
aspects of the relationship between the structure, folding, stability
and functions of proteins. Some proteins are enzymes that promote chemical reactions;
others provide molecular switches that control metabolic and
developmental processes through precise interactions with other
proteins, nucleic acids and other ligands. In two distinct research
programs, we are exploring the chemistry that governs the specificity
and strength of interactions of proteins with other proteins, and
ligands like substrates, inhibitors and nucleic acids.
Some proteins are enzymes that promote chemical reactions;
others provide molecular switches that control metabolic and
developmental processes through precise interactions with other
proteins, nucleic acids and other ligands. In two distinct research
programs, we are exploring the chemistry that governs the specificity
and strength of interactions of proteins with other proteins, and
ligands like substrates, inhibitors and nucleic acids.
 The DnaB interaction domain of DnaG primase.
The DnaB interaction domain of DnaG primase.