
 Go to 2002 RSC Annual Report Index
Go to 2002 RSC Annual Report Index
Biological Chemistry
Protein Synthesis and Evolution
Dr Nicholas Dixon
http://rsc.anu.edu.au/research/dixon.php
Some proteins are enzymes that promote chemical reactions; others provide molecular switches that control metabolic and developmental processes through precise interactions with other proteins, nucleic acids and other ligands. The chemistry that governs the specificity and strength of interactions of proteins with other proteins, and ligands like substrates, inhibitors and nucleic acids, is being explored in two research programs.
The first of these concerns proteins that collaborate to accomplish duplication of the bacterial chromosome prior to cell division. We use these proteins as a model system to study general aspects of protein-protein and protein-nucleic acid interactions. DNA replication is especially useful for this because more than 30 separate proteins act together in a giant nucleoprotein assembly, the replisome, to make a perfect copy of the chromosome. We use molecular genetics to engineer rich sources of the proteins and to produce particular mutant derivatives, and conventional enzymology, DNA synthesis assays and protein chemistry to study protein function. Protein X-ray crystallography, high-field NMR spectroscopy, mass spectroscopy, electron microscopy, and computational methods are used with collaborating laboratories to further understand the structural basis of the functions of the individual proteins and how they interact with each other and with DNA. This year, we have focused our efforts on interaction of the replicative helicase with other replisomal proteins, and on the subunits of Pol III, the DNA polymerase that actually synthesises new DNA chains during replication of chromosomes.
Our other research program has complementary objectives. We are developing a suite of new techniques in protein chemistry, including methods for in vitro evolution of new protein functions, in vitro synthesis of proteins on a preparative scale, library methods for defining precisely the locations of boundaries between distinct folded domains in larger proteins, and stabilisation of small protein domains by end-to-end cyclisation of their polypeptide chains. Used together, these tools will not only help to overcome some of the major bottlenecks in rapid determination of protein structures and functions, thereby increasing the efficiency of worldwide efforts in structural and functional genomics, but they are also used to study fundamental aspects of the relationship between the structure, stability, and function of proteins.
Our most significant breakthroughs this year have been in defining aspects of the active-site chemistry of the proofreading exonuclease subunit of Pol III, in crystallization of the helicase-interaction domain of the DnaG primase, and in development of simple methods for in vitro synthesis and labelling of proteins for structure determination by NMR spectroscopy. Members of the group presented their work at the Lorne Conference on Protein Structure and Function in February, and at ComBio2002 in Sydney in October.
Proteins of DNA Replication
The replisome is made up of several molecular machines that interact physically with each other. One is the replicative DNA polymerase, Pol III, which has ten separate subunits. The e subunit is the proof-reading enzyme that ensures that errors made during DNA replication are immediately recognised and excised before synthesis by the a subunit can continue. A major achievement last year was solution of the X-ray structure of e in a complex with two active-site manganese ions and TMP (one of its products). These data, in conjunction with detailed studies of the activity of the enzyme and modelling of the structure of its complex with DNA, gave us very powerful insights into how e works and why it is such an efficient enzyme. NMR spectroscopic studies of the complex of e with the theta subunit indicate that theta becomes more rigid on binding to e, and solution of the structure of theta in the e.theta complex is nearing completion. The ß subunit is the sliding clamp that prevents Pol III from falling off the DNA template it copies during DNA replication. In collaborative work this year, we have solved the structure of ß by X-ray crystallography at much higher resolution than had been achieved previously. This has given new information about alternate conformations of the molecule in regions that interact with DNA. The DnaB helicase is the molecular motor that separates the two DNA strands in the parental DNA ahead of the polymerase. Like ß, it is a ring-shaped molecule that needs to interact with loading partners to be assembled around very long DNA strands. We have been unable to determine the structure of the hexameric DnaB molecule, but were able to use NMR spectroscopy to solve that of its N-terminal domain. This year, we continued work towards obtaining well-behaved samples of the C-domain, and commenced structural studies on the domain of the DnaG primase that interacts with DnaB at replication forks. Large crystals of this primase domain have now been obtained, and determination of its crystal structure is in progress. (with P.D. Carr, S. Hamdan, M.A. Keniry, P.E. Lilley, K.V. Loscha, M. Mulcair, D.L. Ollis, G. Otting, A.Y. Park, P. Prosselkov, P.M. Schaeffer, and J.M. Carazo [Centro Nacional de Biotecnología, Madrid], C.M. Elvin, G. Wijffels [CSIRO, Brisbane], J.M. Guss [U. Sydney], A. Oakley, M.C.J. Wilce [U. Western Australia])
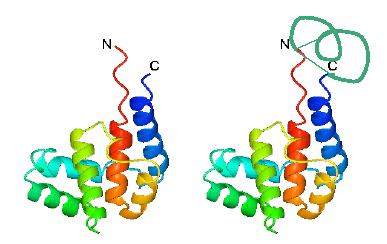
Inteins can be used to join the termini of linear proteins to give circular forms that are much more stable and have other interesting properties (see Williams et al., J. Biol. Chem., 277, 7790-7798, 2002).
New Protein Technologies
Work has continued on development of methods for directed molecular evolution of proteins with new binding specificities. Conditions for selective isolation of binding proteins have now been refined, and selection of their cDNAs from gene libraries can now proceed. Preparative in vitro protein synthesis has been found to be an excellent method for residue-specific incorporation of labelled amino acids into proteins for application of NMR spectroscopic methods to examine ligand binding and for structure determination, and yields of more than 50 proteins in this reaction have been examined. Procedures for generation of high-quality mutant gene libraries for protein domain identification have been devised, and screening gene products will commence early next year. Moreover, our recently-described method for efficient in vivo intein-mediated cyclisation of proteins has been used to generate several new circular proteins and peptides with enhanced stability. Surprisingly, NMR measurements of rates of individual amide proton exchange in one of the cyclic proteins (and a linear analogue) show that exchange from all buried amides occurs at the same rate, and happens when the protein is essentially completely unfolded. In an ongoing collaborative project, electrospray-ionisation mass spectrometry has been used to probe the chemical basis for protein-protein interactions in a complex of two subunits of Pol III. (with M. Headlam, P.E. Lilley, M. Mulcair, G. Otting, K. Ozawa, P. Prosselkov, P.M. Schaeffer, and J.L. Beck, M.M. Shiel [U. Wollongong], J.M. Matthews, N.K. Williams [U. Sydney], M. Ehrenberg [U. Uppsala, Sweden])
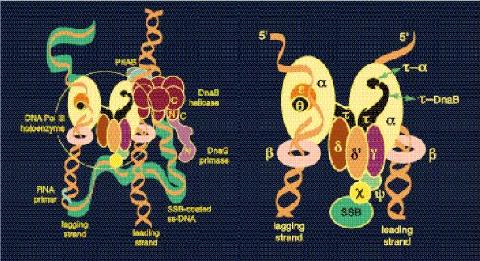
Work over many years has told us a great deal about protein-protein and proten-nucleic acid interactions in the bacterial replisome (left) and among the ten subunits of the DNA polymerase III holoenzyme (right).
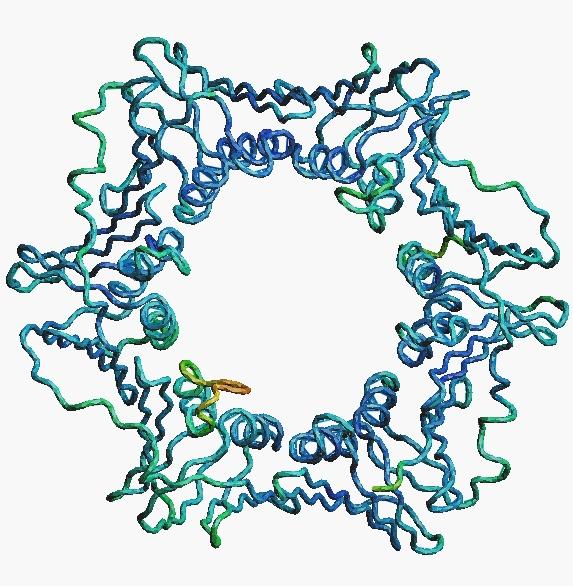
Crystals
grown at RSC have enabled determination of the highest-resolution
X-ray structure of the sliding clamp subunit of DNA polymerase III
holoenzyme. The structure was determined in collaboration with Drs
Aaron Oakley and Matthew Wilce (U. Western Australia).