
 Go to 2002 RSC Annual Report Index
Go to 2002 RSC Annual Report Index
Biological Chemistry
Protein Crystallography and Engineering
Dr David Ollis
http://rsc.anu.edu.au/research/ollis.php
The group works at the interface between chemistry and biology. Our major interest is in working out how proteins function and in how they might be modified for new and useful purposes. The laboratory routinely uses X-ray crystallography to obtain structures that can be used to better understand protein function. Directed evolution is used to produce mutant proteins that frequently have interesting properties that can be utilized in industrial and environmental applications. These mutants can also be analysed using a variety of techniques to further understand the detailed mechanics of protein function.
The structure of an enzyme that degrades organophosphates was solved during the year and directed evolution was used to enhance the catalytic properties of the enzyme. The structure of the e subunit or editing subunit of DNA polymerase III was solved as part of a collaborative study with the Dixon group.
Enzyme Engineering with an Organophosphate Degrading Enzyme
Organophosphate degrading enzyme from Agrobacterium radiobacter (OPDA) is a bacterial enzyme that shows some utility in bioremediation. The protein was initially discovered in the laboratory of John Oakeshott in the Division of Entomology, CSIRO. We have obtained the structure of OPDA using crystallographic techniques and have used directed evolution to probe the catalytic mechanism of the enzyme. (with H. Yang, S. Yu-McLoughlin, P.D. Carr, J. W. Liu)The e Subunit of DNA Polymerase III - the Structure of a Molecular Editor
Proofreading is essential for the faithful reproduction of genetic information. In bacteria, the machinery to carry out this vital checking process is found in the e subunit of DNA polymerase III. The Dixon group expressed an active and soluble fragment that led to the structure that was published this year. (with P.D. Carr, N.E. Dixon, S. Hamdan)
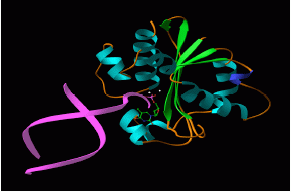
The
X-ray crystal structure of the proofreading subunit of DNA polymerase
III gives important information about how it binds to its DNA
substrate(see Hamdan et al., Structure, 10, 535-546,
2002).
The ß Subunit of the IL-5 Receptor - Identifying the Interaction Site
IL-5 is a regulator of growth, differentiation, and activation of the white blood cell eosinophils. These cells are of major importance in the body's response to invasion by parasites and asthma inducing aeroallergens. Last year the structure of the ß receptor interleukin-5 (IL-5) was published. Efforts to exploit the new structure have proceeded this year. (with P.D. Carr, J. Murphy, and I.G. Young [JCSMR, ANU])
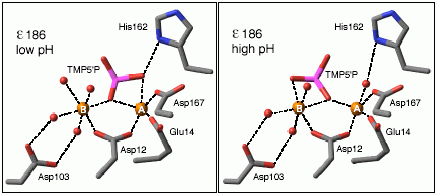
Structures of thymidine monophosphate bound at the active site of the proofreading subunit of DNA polymerase III at two different pH values give important insights into how the enzyme works (see Hamdan et al., Structure, 10, 535-546, 2002; and Biochemistry, 41, 5266-5275, 2002).